This research was originally published at ACM BCB 2014. The publication can be downloaded here.
ABSTRACT
The Super Threaded Reference-Free Alignment-Free N-sequence Decoder (Strand) is a highly parallel technique for the learning and classication of gene sequence data into any number of associated categories or gene sequence taxonomies. Current methods, including the state-of-the-art sequence classication method RDP, balance performance by using a shorter word length. Strand in contrast uses a much longer word length, and does so eciently by implementing a Divide and Conquer algorithm leveraging MapReduce style processing and locality sensitive hashing. Strand is able to learn gene sequence taxonomies and classify new sequences approximately 20 times faster than the RDP classifierer while still achieving comparable accuracy results. This paper compares the accuracy and performance characteristics of Strand against RDP using 16S rRNA sequence data from the RDP training dataset and the Greengenes sequence repository.
1. INTRODUCTION
Many popular gene sequence analysis tools are based on the idea of pairwise sequence alignment [16, 19]. Sequence alignment finds the least costly transformation of one sequence into another using insertions, deletions, and replacements. This method is related to the well know distance metric called Levenshtein or edit distance. However, finding the optimal pairwise alignment is computationally expensive and requires dynamic programming.
Gene sequence words are sub-sequences of a given length. In addition to words they are often also referred to as k-mers or n-grams, where k and n represent the word length. Words are extracted from individual gene sequences and used for similarity estimations between two or more gene sequences [21]. Methods like BLAST [1] were developed for searching large sequence databases. Such methods search for seed words first and then expand matches. These so called alignment-free methods [21] are based on gene sequence word counts and have become increasingly popular since the computationally expensive sequence alignment method is avoided. One of the most successful word-based methods is the RDP classifier [22], a naive Bayesian classifier widely used for organism classification based on 16S rRNA gene sequence data.
Numerous methods for the extraction, retention, and matching of word collections from sequence data have been studied. Some of these methods include: 12-mer collections with the compression of 4 nucleotides per byte using byte-wise searching [1], sorting of k-mer collections for the optimized processing of shorter matches within similar sequences [11], modification of the edit distance calculation to include only removals (maximal matches) in order to perform distance calculations in linear time [20], and the use of locality sensitive hashing for inexact matching of longer k-mers [5].
This research combines the following three primary contributions in a novel and innovative way to achieve the results presented:
- Jaccard similarity is used as an approximation for edit distance when determining the similarities and differences between gene sequence data.
- A form of locality sensitive hashing called minhashing is used to rapidly process much longer word lengths for enhanced accuracy. Minhashing allows us to estimate Jaccard similarity without computing and storing information for all possible words extracted from a gene sequence. Instead, we use the intersection of the minhash signatures produced during the minhashing process to quickly calculate an accurate approximation of the Jaccard similarity between sequences and known taxonomy categories.
- A MapReduce style parallel pipeline is used to simultaneously identify unique gene sequence words, minhash each word generating minhash signatures, and intersect minhash signatures to estimate Jaccard similarity for highly accurate and efficient identification of gene sequence taxonomies.
Buhler [5] previously used locality sensitive hashing to allow for inexact matches between longer words of a predefined length. We use locality sensitive hashing in a very different way as a selection strategy for performing exact word matching when the number of possible words becomes much too large to store. For example, with an alphabet of 4 symbols, the number of unique words of length 60 is 460 which is already more than 1036 distinct words. The RDP classifier utilizes a fixed word length of only 8 bases to perform its taxonomy classification processing making the total possible number of unique words (i.e., features for the classifier) only 4^8 = 65,536 words [22].
Strand is able to very rapidly classify sequences while still taking advantage of the increased accuracy provided by extracting longer words. Using the much larger possible feature space provided by a longer word length combined with locality sensitive hashing to reduce memory requirements, Strand achieves classification accuracy similar to RDP in processing times which are magnitudes of order faster. All stages of Strand processing are highly parallelized, concurrently mapping all identified words from a gene sequence and reducing mapped words into minhash signatures simultaneously. The unique relationship of Jaccard similarity between sets and locality sensitive hashing [17] allows minhashing to occur during learning, storing only a predetermined number of minimum hash values in place of all words extracted from the gene sequences in the learning set. This process reduces the amount of memory used during learning and classification to a manageable amount.
2. BACKGROUND
This paper combines the three concepts of longer length word extraction, minhashing, and multicore MapReduce style processing in a novel way that produces superior gene sequence classification results. The following sections briefly describe the background material relevant to this research.
2.1 Word Extraction
The general concept of k-mers or words was originally defined as n-grams during 1948 in an information theoretic context [18] as a sub-sequence of n consecutive symbols. We will use the terms words or k-mers in this paper to refer to n-grams created from a gene sequence. Over the past twenty years, numerous methods utilizing words for gene sequence comparison and classification have been presented [21]. These methods are typically much faster than alignment-based methods and are often called alignment-free methods. The most common method for word extraction uses a sliding window of a fixed size. Once the word length k is defined, the sliding window moves from left to right across the gene sequence data producing each word by capturing k consecutive bases from the sequence.
Strand performs word extraction using lock-free data structures to identify unique gene sequence words. This method is similar to other highly parallel word counting tools such as Jellysh [14]. Traditionally, computationally expensive lock objects are used in parallel programs to synchronize thread-level access to a shared resource. Each thread must either acquire the lock object or block until it becomes available prior to entering critical sections of code. Lock-free data structures avoid the overhead associated with locking by making use of low-level atomic read/write transactions and other lock-free programming techniques.
2.2 Minhashing
In word-based sequence comparison, sequences are often considered to be sets of words. A form of locality sensitive hashing called minhashing uses a family of random hash functions to generate a minhash signature for each set. Each hash function used in the family of n hash functions implement a unique permutation function, imposing an order on the set to be minhashed. Choosing the element with the minimal hash value from each of the n permutations of the set results in a signature of n elements. Typically the original set is several magnitudes larger than n resulting in a significant reduction of the memory required for storage. From these signatures an estimate of the Jaccard similarity between two sets can be calculated [2, 17]. Minhashing has been successfully applied in numerous applications including estimating similarity between images [3] and documents [2], document clustering on the internet [4], image retrieval [8], detecting video copies [6], and relevant news recommendations [13].
In this paper we apply minhashing to estimate the similarity between sequences which have been transformed into very large sets of words.
2.3 MapReduce Style Processing
MapReduce style programs break algorithms down into map and reduce steps which represent independent units of work that can be executed using parallel processing [7, 10]. Initially, input data is split into many pieces and provided to multiple instances of the mapping functions executing in parallel. The result of mapping is a key-value pair including an aggregation key and its associated value or values. The key-value pairs are redistributed using the aggregation key and then processed in parallel by multiple instances of the reduce function producing an intermediary or final result.
MapReduce is highly scalable and has been used by large companies such as Google and Yahoo! to successfully manage rapid growth and extremely massive data processing tasks [15]. Over the past few years, MapReduce processing has been proposed for use in many areas including: analyzing gene sequencing data [15], machine learning on multiple cores [12], and highly fault tolerant data processing systems [7, 9].
Strand uses MapReduce style processing to quickly map gene sequence data into words while simultaneously reducing the mapped words from multiple sequences into their appropriate corresponding minhash signatures.
3. LEARNING CATEGORY SIGNATURES
Figure 1 illustrates a high-level process of the Strand MapReduce pipeline including both the mapping of gene sequence data into words, the reduction of words into minimum hash values, and finally, the last reduce step which organizes the minhash signatures by category. In the following we will describe each stage in detail.
3.1 Mapping Sequences into Words
The input data are sequences with associated categories.
DEFINITION 1 (Sequence). Let S be a single input sequence, a sequence of |S| symbols from alphabet Σ = {A, C, G, T}.
DEFINITION 2 (Category). Let C be the set of all L known taxonomic categories and cl ∈ C be a single category where l = {1, 2, … , L}. Each sequence S is assigned a unique true category cl ∈ C.
The goal of mapping sequences into words is to create for each sequence a word profile.
DEFINITION 3 (Sequence Word Profile). Let S denote the word profile of sequence S, i.e., the set of all words sj ∈ S, j = {1, 2, … , |S|}, with length k extracted from sequence S.
3.2 Creating Sequence Minhash Signatures
As words are produced, minhashing operations are also performed simultaneously in parallel to create minhash signatures.
DEFINITION 4 (Sequence Minhash Signature). Minhashing (min-wise locality sensitive hashing) applies a family of random hashing functions h1, h2, h3,…hk to the input sequence word profile S to produce k independent random permutations and then chooses the element with the minimal hash value for each. We define the minhash function:
Min-wise locality sensitive hashing operations are performed in parallel and continually consume all randomly selected word hashes for each processed sequence. The minhash signature’s length is predetermined by the number of random hashing functions used during minhash processing, and the minhash signature length impacts processing time and overall classification accuracy. Processes using more hashing functions (i.e., longer minhash signatures) have been proven to produce more accurate Jaccard estimations [17]. However, careful consideration must be given to the tradeoff between the minhash signature’s impact on performance and Jaccard estimation accuracy.
A thread-safe minhash signature collection contains one minhash signature for each unique input sequence. During minhash processing, all hash values produced for each sequence’s unique set of word hashes are compared to the sequence’s current minhash signature values. The minimum hash values across all unique words for each sequence and each unique hashing function are then retained within each sequence’s final minhash signature. In applications where the similarity calculation incorporates word length or the frequency of each minimum hash value, the length and frequency for any word resulting in a particular minimum hash value can also be contained as additional properties within each minhash signature’s values. However, lengths are not retained within the Strand data structure during training since they can quickly be determined during any subsequent classification’s minhashing process. In some cases, the total number of hashing functions and overall minhashing operations can be reduced by saving the n smallest hash values generated from each individual hashing function. For instance, the number of hashing operations can be cut in half by retaining the 2 smallest values produced from each unique hashing function.
3.3 Reducing Sequence Minhash Signatures into Category Signatures
Next we discuss how to represent an entire category as a signature built from the minhash signatures of all sequences in the category.
DEFINITION 5 (Category Minhash Signature). We define the category minhash signature of category cl ∈ C as the union of the sequence minhash signatures of all sequences assigned to category cl:
where the union is calculated for each minhash hashing function separately.
The Strand data structure actually represents an array containing one data structure for each unique hashing function used (see Figure 1). Since this structure is keyed using minhash values, hash function partitions must exist to separate the minhash values produced by each unique hashing function. Within each individual hash function partition, a collection of key-value pairs (kvp) exists which contains the minhash value as a key and then a second nested collection of categorical key-value pairs for each value. The nested collection of kvp-values contains all category numbers and associated frequencies (when required) that have been encountered for a particular minhash value. In practice however, minhash values seldom appear to be associated with more than one taxonomy category which drastically reduces the opportunity for imbalance between categories, especially when minhash value frequencies are not used within the classification similarity function.
During learning, minimum hash values for each unique hashing function are retained within the array of nested categorical key-value pair collections and partitioned by each unique hashing function. Each hash function’s collection contains all unique minimum hash values, their associated taxonomies, and optional frequencies. Using the Strand data structure, minhash signatures for each input data sequence can quickly be compared to all minimum hash values associated with each known taxonomy including the taxonomy frequency (when utilized) during classification. During training, each value in the input data sequence’s minhash signature is reduced into the Strand data structure by either creating a new entry or adding additional taxonomy categories to an existing entry’s nested categorical key-value pair collection.
All results presented in this research were achieved using only a binary classication similarity function. This approach produced optimal performance while still achieving comparable accuracy results when benchmarked against the current top performer in this domain.
4. CLASSIFICATION PROCESS
The MapReduce style architecture used for learning and classification are very similar. While the process of mapping words into minhash signatures is identical, the reduce function now instead of creating category signatures creates classification scores.
The word profiles of sequences are the set of words contained within the sequences. A common way to calculate the similarity between sets is the Jaccard index. The Jaccard index between two sequence word profiles S1 and S2 is defined as:
However, after minhashing we only have the sequence minhash signatures M1 = minhash(S1) and M2 = minhash(S2) representing the two sequences. Fortunately, minhashing [17] allows us to efficiently estimate the Jaccard index using only the minhash signatures:
where the intersection is taken hash-wise, i.e., how many minhash values agree between the two signatures.
Next, we discuss scoring the similarity between a sequence minhash signature and the category minhash signatures used for classification. Category signatures are not restricted to k values since they are created using the unique minhash values of all sequence minhash signatures belonging to the category. This is why we do not directly estimate the Jaccard index, but dene a similarity measure based on the number of collisions between the minhash values in the sequence signature and the category signature.
DEFINITION 6 (Minhash Category Collision). We define the Minhash Category Collision between a sequence S represented by the minhash signature M and a category signature C as: 
We calculate MCC for each category and classify the sequence to the category resulting in the largest category collision count.
Many other more sophisticated approaches to score sequences are possible. These are left for future research.
5. RESULTS
In this section we report on a set of initial experiments. First, we compare dierent word sizes and numbers of sequence signature lengths (i.e., the number of hashing functions used for minhashing). Then we compare Strand with RDP using two different data sets.
The data sets we use are all 16S rRNA data sets. The RDP classifier raw training set was obtained from the RDP download page. It contains 9,217 sequences. The second data set we use is extracted from the Greengenes database. We randomly selected 150,000 unaligned sequences with complete taxonomic information for our experiments. We used for all experiments 10-fold cross-validation. During 10-fold cross-validation, the entire training file is randomly shuffled and then divided into ten equally sized folds or segments. While nine folds are learned, one fold is held out for classification testing. This process is repeated until all ten folds have been held out and classified against.
The experiments were performed on a Windows 8 (64-bit) machine with a 2.7 Ghz Intel i7-4800MQ quad core CPU and 28 GB of main memory installed. For the RDP classier we used version 2.5 (rdp-classifier-2.5.jar), and we implemented Strand in C#.
5.1 Choosing Word Size and Signature Length
Both, the used word size and the length of the signature need to be specified for Strand. We expect both parameters to have an impact on classification accuracy, space, and run time. While it is clear that with increasing signature lengths also the time needed to compute sequence signatures and the space needed to store signatures increases, the impact of word size is not so clear. In the following we will empirically find good values for both parameters.
To look at the impact of the word length, we set the signature size (i.e., number of hash functions used for minhashing) to 300. This was empirically found to be a reasonable value. Next we perform 10-fold cross-validation on the RDP training data for different word lengths ranging from 8 bases to 200 bases. The average accuracy of Genus prediction and overall prediction (average accuracy over all phylogenetic ranks) depending on the word length is shown in Figure 2. We see that accuracy increases with word length till the word length reaches 60 bases and then starts to fall quickly at lengths larger than 100 bases. This shows that the optimal word length for the used 16S rRNA is around 60 bases.
Next, we look at the impact of sequence signature length. We use a xed word size of 60 and perform again 10-fold cross-validation on the RDP training data set using signature lengths from 50 to 500. Figure 3 shows the impact of signature length. Accuracy initially increases with signature length, but flattens at about 300. Since an increased signature length directly increases run time (more hashes need to be calculated) and storage, we conclude that 300 is the optimal size for the used 16S rRNA data, but signature lengths of 200 or even 100 also provide good accuracy at lower computational cost.
5.2 Comparison of Strand and RDP on the RDP Training Data
In this section we compare Strand and RDP in terms of run time and accuracy. For the comparison we use again 10-fold cross-validation on the RDP training data set. Table 1 shows the run time for learning and classification using Strand and RDP. While the learning times for Strand are approximately 30% faster, Strand classifies sequences almost 20 times faster than RDP. Strand trained with 8,296 sequences averaging around 23 seconds per fold while RDP averaged around 33 seconds on the same 8,296 training sequences. During 10-fold cross-validation, Strand was able to classify 921 sequences averaging 3 seconds per fold while RDP’s average classification time was 59 seconds per fold. This is substantial since classification occurs much more frequently than training in a typical application. Since Strand uses longer words during training and classification, no bootstrap sampling or consensus is used to determine taxonomy assignments. Strand greatly increases classification speeds when compared to RDP by combining this factor with a highly parallel MapReduce style processing pipeline.
An accuracy comparison between Strand and RDP is shown in Table 2. In cross-validation it is possible that we have a sequence with a Genus in the test set for which we have no sequence in the learning set. We exclude such sequences from the results since we cannot predict a Genus which we have not encountered during learning. Strand achieves similar overall accuracy to RDP, however, as we saw above in a fraction of the time.
5.3 Comparison of Strand and RDP on the Greengenes Data
Here we compare Strand and RDP on a sample of 150,000 sequences from the Greengenes project. While the RDP training set is relatively small and well curated to create a good classier, these sequences will contain more variation. To analyze the impact of data set size, we hold 25,000 sequences back for testing and then use incrementally increased training set sizes from 25,000 to 125,000 in increments of 25,000 sequences. For Strand we use a word size of 60 and a sequence signature length of 100.
Figure 4 shows the classication accuracy of Strand and RDP using an increasingly larger training set. Strand has slightly higher accuracy. Accuracy increases for both classifiers with training set size. However, it is interesting that after 100,000 sequences, the accuracy starts to drop with a significantly steeper drop for RDP.
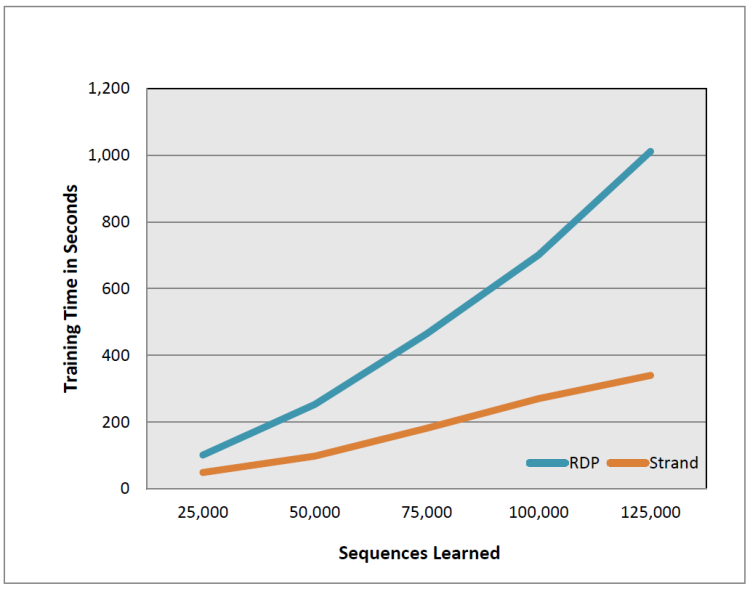
Figure 5 compares the time needed to train the classifiers with increasing training set size. During training, Strand execution times consistently outperform RDP with training time deltas further widening as input training volumes increase. In Figure 5 RDP training times increase rapidly as the training set size increases. Strand training times increase at a rate closer to linear. When training against 125,000 Greengenes sequences, Strand completes training in 5:39 (mm:ss) while RDP takes 16:50 (mm:ss). The classification time does not vary much with the training set size. Strand’s average classication time for the 25,000 sequences is 1:41 (mm:ss) while the average time for RDP is 20:15 (mm:ss).
Finally, we look at memory requirements for Strand. Since we use a word size of 60 bases, there exist 4^60 = 10^36 unique words. For RDP’s word size of 8 there are only 4^8 = 65,536 unique words. Strand deals with the huge amount of possible words using minhashing and adding only a small signature for each sequence to the class signature. This means that the Strand data structure will continue to grow as the volume of training data increases. The overall space consumption characteristics of Strand are directly related to the selected word size, the minhash signature length, and the amount of sequences learned. Figure 6 shows the percentage of unique signature entries (minhash values) stored relative to the number of words processed. With increasing training set size the fraction of retained entries falls from 2% at 25,000 sequences to just above 1% at 125,000 sequences. This characteristic is attributed to the fact that many sequences share words. In total, Strand stores for 125,000 sequences 1.7 million entries which is more than the 65,536 entries stored by RDP, but easily ts in less than 1 GB of main memory which is typically in most modern smart phones.
6. CONCLUSION
In this paper we have introduced a novel word-based sequence classification scheme that utilizes large word sizes. A highly parallel MapReduce style pipeline is used to simultaneously map gene sequence input data into words, map words into word hashes, reduce all word hashes within a single sequence into a minimum hash signature, and then populates a data structure with category minhash signatures which can be used for rapid classification. Experiments show that for 16S rRNA a word size of 60 bases and a sequence minhash signature length of 300 produce the best classification accuracy. Compared to RDP, Strand provides comparable accuracy while performing classification 20 times as fast.
Acknowledgements
The systems and methods described in this publication are protected by the following US patent applications:
- Jake Drew. 2014. Collaborative Analytics Map Reduction Classification Learning Systems and Methods. (Feb. 2013). Patent Application No. 14/169,689, Filed February 6th., 2013.
- Jake Drew, Michael Hahsler, Tyler Moore. 2014. System and Method for Machine Learning and Classifying Data. (May 2013). Patent Application No. 14/283,031, Filed May 20th., 2013.
7. REFERENCES
- S. F. Altschul, W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. Basic local alignment search tool. Journal of Molecular Biology, 215(3):403-410, 1990.
- A. Z. Broder. On the resemblance and containment of documents. In Compression and Complexity of Sequences 1997. Proceedings, pages 21-29. IEEE, 1997.
- A. Z. Broder, M. Charikar, A. M. Frieze, and M. Mitzenmacher. Min-wise independent permutations. In Proceedings of the thirtieth annual ACM symposium on Theory of computing, pages 327-336. ACM, 1998.
- A. Z. Broder, S. C. Glassman, M. S. Manasse, and G. Zweig. Syntactic clustering of the web. Computer Networks and ISDN Systems, 29(8):1157-1166, 1997.
- J. Buhler. Ecient large-scale sequence comparison by locality-sensitive hashing. Bioinformatics, 17(5):419-428, 2001.
- C.-Y. Chiu, H.-M. Wang, and C.-S. Chen. Fast min-hashing indexing and robust spatio-temporal matching for detecting video copies. ACM Transactions on Multimedia Computing, Communications, and Applications (TOMCCAP), 6(2):10, 2010.
- C. T. Chu, S. K. Kim, Y. A. Lin, Y. Yu, G. R. Bradski, A. Y. Ng, and K. Olukotun. Map-Reduce for Machine Learning on Multicore. In B. Scholkopf, J. C. Platt, and T. Homan, editors, NIPS, pages 281-288. MIT Press, 2006.
- O. Chum, M. Perdoch, and J. Matas. Geometric min-hashing: Finding a (thick) needle in a haystack. In Computer Vision and Pattern Recognition, 2009. CVPR 2009. IEEE Conference on, pages 17-24. IEEE, 2009.
- J. Dean and S. Ghemawat. Mapreduce: A flexible data processing tool. Communications of the ACM, 53(1):72-77, 2010.
- J. Drew. Mapreduce: Map reduction strategies using C#, 2013.
- R. C. Edgar. Search and clustering orders of magnitude faster than BLAST. Bioinformatics (Oxford, England), 26(19):2460-1, 2010.
- D. Keco and A. Subasi. Parallelization of genetic algorithms using hadoop map/reduce. South East Europe Journal of Soft Computing, 1(2), 2012.
- L. Li, D. Wang, T. Li, D. Knox, and B. Padmanabhan. Scene: A scalable two-stage personalized news recommendation system. In ACM Conference on Information Retrieval (SIGIR), 2011.
- G. Marcais and C. Kingsford. A fast, lock-free approach for ecient parallel counting of occurrences of k-mers. Bioinformatics, 27(6):764-770, 2011.
- A. McKenna, M. Hanna, E. Banks, A. Sivachenko, K. Cibulskis, A. Kernytsky, K. Garimella, D. Altshuler, S. Gabriel, M. Daly, et al. The genome analysis toolkit: A mapreduce framework for analyzing next-generation dna sequencing data. Genome research, 20(9):1297-1303, 2010.
- S. B. Needleman and C. D. Wunsch. A general method applicable to the search for similarities in the amino acid sequence of two proteins. Journal of Molecular Biology, 48(3):443-453, 1970.
- A. Rajaraman and J. Ullman. Mining of Massive Datasets. Mining of Massive Datasets. Cambridge University Press, 2012.
- C. E. Shannon. A mathematical theory of communication. The Bell Systems Technical Journal, 27:379-423, 1948.
- T. F. Smith and M. S. Waterman. Identication of common molecular subsequences. Journal of Molecular Biology, 147(1):195-197, March 1981.
- E. Ukkonen. Approximate string-matching with q-grams and maximal matches. Theoretical Computer Science, 92(205):191{211, 1992.
- S. Vinga and J. Almeida. Alignment-free sequence comparison | A review. Bioinformatics, 19(4):513-523, 2003.
- Q. Wang, G. M. Garrity, J. M. Tiedje, and J. R. Cole. Naive bayesian classier for rapid assignment of RNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology, 73(16):5261-5267,2007.